U. Deva Priyakumar, PhD
Professor, CCNSB, IIIT-Hyderabad
Ph.D (Pondicherry University)
Research Areas:
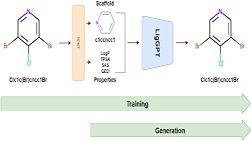
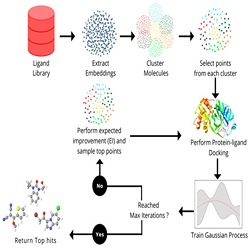
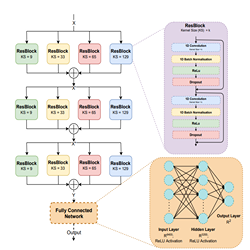
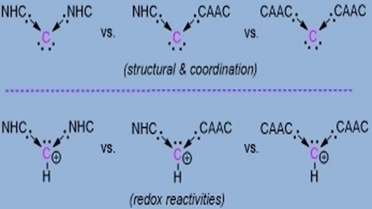
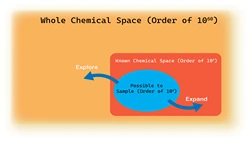
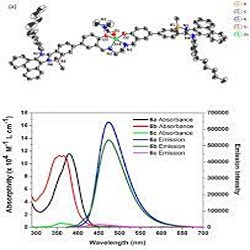
Computational chemistry to study chemical molecules and reaction mechanisms, biomolecular simulations to investigate DNA, RNA, proteins and protein interactions and computer aided drug design
Email:deva@iiit.ac.in
Address:
International Institute of Information Technology
Gachibowli
Hyderabad - 500 032
India
Phone: (91) (40) 6653 1000 Ext: 1161